
Journal of Animal Research: v.10 n.1, p. 127-131. February 2020
DOI: 10.30954/2277-940X.01.2020.18
Occurrence of Methicillin Resistant Staphylococcus aureus from Bovine Raw Milk in Chennai
ABSTRACT
Staphylococcus aureus and Methicillin-Resistant Staphylococcus aureus (MRSA) presence in milk is of significant importance as they are often involved in food borne intoxications. Effective surveillance and detection are the most important tools in outbreak investigation and control. Our study was designed to ascertain the occurrence Staphylococcus aureus and Methicillin-Resistant S. aureus (MRSA) from raw milk collected from apparently healthy cattle in Chennai city, Tamil Nadu, India. A total of 89 (89/258; 34.49%) positive isolates of Staphylococcus aureus obtained from 258 raw milk samples screened. Of which 24 (24/258; 9.3%) isolates positive for MRSA. Studying the antimicrobial resistance and its patterns in foods of animal origin will be of immense help to various regulatory agencies and authorities to devise and implement control measures and to note the compliance of our foods with the prescribed standards.
Keywords: MRSA, Staphylococcus aureus, Prevalence, Oxacillin Resistance, Raw milk
Staphylococcus aureus is an opportunistic, Gram-positive cocci and they are catalase and coagulase positive microbes implicated in various hospital acquired and food borne outbreaks. Niche of this bacterium varies from environmental samples to skin and mucosa of both animals and human (Tong et al., 2015). In dairy industry, S. aureus is the most prevalent and economically significant important pathogen (Katsuda et al., 2005). The organism enters milk either through direct contamination by infection or normal inhabitation and/or indirectly by external environmental contamination or through animal handlers (Jorgensen et al., 2005). S. aureus contamination of milk with enterotoxigenic staphylococcal enterotoxins (SEs) production leads to intoxication and it is often associated with symptoms like vomition and diarrhoea (Veras et al., 2008). The major problem with food borne S. aureus is that,it’s a prime pathogen involved in antimicrobial resistance with the potential of transferring between animals and humans.
In 1959, methicillin was introduced as the first semisynthetic penicillin to combat penicillinase-producing S. aureus penicillin to combat penicillinase-producing S. aureus which were found resistant to penicillin (Livermore, 2000). The resistance to methicillin was noticed within a year of introduction and it was postulated that overproduction of PBP2a, penicillin-binding protein which has less affinity for beta-lactam antibiotics (Chambers, 1997). The methicillin-resistant S. aureus (MRSA) is a rampant public health issue with human health concern. MRSA harbours staphylococcal cassette chromosome mec (SCCmec) complex which aids in resistance to methicillin. According to the Center for Disease Control, two out of every 100 carriers of S. aureus are carriers of MRSA (CDC, 2017). The mecA gene, a structural determinant encoding PBP2a, is therefore considered to be a useful molecular marker of putative methicillin resistance in S. aureus. The MRSA has clonal derivatives from healthcare associated infections (HA-MRSA) worldwide (Köck et al., 2010). Similarly, community (CA-MRSA for community acquired MRSA) (David and Daum, 2010) and in livestock (LA-MRSA for livestock associated MRSA) (Nemati et al., 2009). Hence, the present study was designed to envisage the occurrence of methicillin-resistant S. aureus (MRSA) from raw milk collected from apparently health cattle’s in Chennai city, Tamil Nadu, India.
How to cite this article: Deepak, S.J., Porteen, K., Elango, A., Kumar, TMAS., Babu, R.N., Sureshkannan, S. and Ruban, S.W. (2020). Occurrence of methicillin resistant Staphylococcus aureus from bovine raw milk in Chennai. J. Anim. Res. 10(1): 127-131.
MATERIALS AND METHODS
Study design and sampling
A cross-sectional study was designed to analyse raw milk collected between October 2016 to May 2019. The milk samples were collected from various farms locations of Chennai city, Tamil Nadu, India. Purposive sampling technique was employed. A total of 258 raw milk samples were collected aseptically, labelled and transported in ice box to laboratory for bacterial isolation and characterization. Processing of samples or carried on same day or kept in refrigerator at 4°C until microbial identification.
Isolation and confirmation of S.aureus
The milk samples were enriched in brain heart infusion broth containing 5-7% NaCl. The sample were diluted at 1:10 dilution for enrichment in broth and incubated at 35-37°C for 18-24 h. The enriched samples plated on Mannitol Salt Agar (MSA)incubated for 24 h at 35-37°C. The characteristic appearance of golden yellow colour colonies was considered to be presumptive S. aureus. The organism was demonstrated by Gram’s stained smears which show Gram-positive cocci that occurred in grape like clusters. Further confirmation was done by positive reaction with catalase test (3% hydrogen peroxide), slide coagulase test, latex agglutination test and confirmed by nuc gene based PCR assay.
DNA Preparation and PCR assay
The DNA was extracted by hot boiling and snapchill method is used further PCR based studies. The supernatant was used as DNA template for PCR reaction mixture. The polymerase chain reaction was performed in a reaction mixture of 25 µL of final volume approximately containing 50 ng of template DNA, 12.5 µL of Taq DNA polymerase 2X Master mix (Amplicon III, Denmark), 1 µL of (10 pmol) each primer (Sigma Aldrich, India) and made up volume by nuclease free water. The PCR was performed in an A200 Gradient Thermal Cycler (LongGene®, Hangzhou, China). The gel was visualized using Imagecapture software in BioRad Gel documenter (Bio rad).
Multiplex PCR for confirmation and detection of MRSA
The nuc gene (thermonuclease) (Hegde et al. 2013) and mecA complex (methicillin A gene) (Ryffel, 1990) with oligonucleotide primers and cycling conditions used are given in Table 1. The multiplex PCR was designed using established primers were used for simultaneous detection of nuc gene and mecAgene from S. aureus isolates from test milk sample.
Phenotypic identification of MRSA
Antimicrobial susceptibility test, by disk diffusion method was performed for all S. aureus isolates as per the protocol of Clinical and laboratory standards institute, 2017 (CLSI, 2017). The turbidity of test isolates was adjusted to 0.5 McFarland standard units. The bacterial suspension was spread onto Muller–Hinton agar (Himedia, India) with the sterile swab and left at room temperature to dry. Following cefoxitin (CX-30) disc was placed on bacterial lawn prepared. The plates were incubated at 37°C for 24 h. The zone of inhibition around the disks were measured and classified as susceptible or resistant according to the CLSI, 2017.
RESULTS AND DISCUSSION
In the present study, a total of 89 (89/258; 34.49%) positive isolates of Staphylococcus aureus obtained from 258 raw milk samples screened. The isolates were confirmed by various specific biochemical tests and nuc gene based PCR assay. By staining the isolates exhibited clusters of grape appearance and gram-positive cocci with the ability to ferment mannitol and produce golden yellow colour colonies on Mannitol salt agar (MSA). Further biochemical characterization showed positive reaction with catalase on 3% hydrogen peroxide and slide coagulase test using rabbit coagulase plasma (Himedia, India). The molecular confirmation of S. aureus isolates with nuc gene (Thermonuclease) amplified a 188 bp specific amplicon product. Sudhanthiramani et al. (2015) reported that prevalence of 39.09% (43/110) coagulase-positive S. aureus from the milk samples which is in accordance with our study, whereas Bhati et al. (2018) reported 107/197 (54.31%) S. aureus isolates confirmed from mastitic milk samples, udder and milkers’ hands by 23s rRNA based PCR in Rajasthan, India. Akriti et al. (2019) documents the prevalence of S. aureus as 66.66% (40/60) in the cattle milk samples collected from Vallabhnagar tehsil of Udaipur district. Sarkar et al. (2014) recorded 74.5% prevalence of S. aureus in milk samples, Haryana, India. Gundogan and Avci, (2014) reported that 56% of prevalence of S. aureus in raw milk and dairy product in Turkey. Variation in the occurrence of S. aureus may be attributed to various factors like sampling design, study location and methodology adopted.
Table 1: The list of primers and cycling condition used in the present study

The phenotypic method for identification of methicillin resistant (oxacillin resistant) S. aureus (MRSA) utilizing cefoxitin antimicrobial disc is recommended as it is an effective identification marker of mecA presence as per CLSI, 2017. Genotypic identification of methicillin resistant S. aureus (MRSA) confirmation is based on presence on mecA gene is recommended by various researchers (Palavecino, 2014). The cefoxitin has higher sensitivity and specificity over oxacillin test for detection of mecA from MRSA and MSSA strains (Elhaig and Selim, 2015). The multiplex PCR was designed for identifying both nuc gene based species specific S. aureus with an amplicon size 188 bp and mecA gene based MRSA with an amplicon size of 310 bpwas amplified from the genomic DNA (Fig. 1).
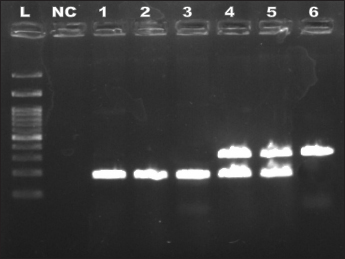
Fig. 1: The PCR products Gel image representing the products of multiplex PCR developed
L: DNA ladder 100kb; NC-Negative control; 1-3- nuc positive SA (181bp); 4-5- mecA positive MRSA (181bp & 310bp) and 6- mecA positive other Staphlyococcus spp. (310bp)
A total of 24 (24/258; 9.3%) isolates positive for MRSA on both phenotypic and genotypic methods respectively. Kriegeskorte et al. (2012) reported that phenotypic methods are accurate identification of MRSA isolates along with genotypic identification of mecA, mecC, and other Staphylococcal chromosomal cassette mec. Riva et al. (2015) found that the prevalence of 9.1% S. aureus in raw milk and the 20% of which are MRSA in Italy which is similar to our results. In Germany, Kreausukon et al. (2012) reported that 4.4% are MRSA in bulk tank milk analysed. Normanno et al. (2007) reported 17% S. aureus in milk and dairy products and 3.75% were MRSA. The presence of MRSA from milk samples of apparently healthy animals is an important issue in dairy food production. The conditions like improper hygiene and poor management practices on before and during milking will aid in the contamination of milk with S. aureus which may eventually leads to emergence of antibiotic resistant strains.
CONCLUSION
In the present study we have found that a total of 89 (89/258; 34.49%) positive isolates of Staphylococcus aureus and 24 (24/258; 9.3%) isolates positive for MRSA on both phenotypic and genotypic methods. The multiplex PCR assay employed was identified as easy, no-ambiguous and rapid in identification of S. aureus and MRSA, respectively. Our study identified the presence of MRSA strains which needs due attention to curtail the spread of this potential public health risk associated antibiotic resistant strains among animals and human.
REFERENCES
Akriti, Diwakar. and Gaurav, A. 2019. Prevalence and antibiotic resistance pattern of Staphylococcus aureus of dairy origin from Udaipur (Rajasthan) region. J. Entomol. Zool. Stud.,7(4): 1143-1145.
Bhati, T., Gaurav, K., Khichar, V. and Kataria, A.K. 2018. Prevalence of Staphylococcus aureus isolated from mastitic milk, udder Surfaces and milkers’ hands from different farms in Bikaner, Rajasthan. J. Anim. Res., 8(5): 867.
Centers for Disease Control and Prevention (CDC).2017. Surveillance for foodborne disease outbreaks, United States, 2015, Annual report. Atlanta, Georgia: US Department of health and human Services, CDC, 2017
Chambers, H.F. 1997. Methicillin resistance in staphylococci: molecular and biochemical basis and clinical implications. Clin. Microbiol. Rev., 10(4): 781-791.
CLSI.2017. Performance standards for antimicrobial susceptibility testing; twenty-fourth informational supplement. Wayne: CLSI document M100-S26; 2017.
David, M.Z. and Daum, R.S. 2010. Community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin. Microbiol. Rev., 23(3): 616-687.
Elhaig, M.M. and Selim, A. 2015. Molecular and bacteriological investigation of subclinical mastitis caused by Staphylococcus aureus and Streptococcus agalactiae in domestic bovids from Ismailia, Egypt. Trop. Anim. Health. Pro., 47(2): 271-276.
Elizabeth L. Palavecino. 2014. Chapter 3: Rapid Methods for Detection of MRSA in Clinical Specimens. YinduoJi (ed.), Methicillin-Resistant Staphylococcus aureus (MRSA) Protocols, Methods in Molecular Biology, vol. 1085, DOI 10.1007/978-1-62703-664-1_3, Springer Science+Business Media, LLC 2014
Gundogan, N. and Avci, E. 2014. Occurrence and antibiotic resistance of Escherichia coli, Staphylococcus aureus and Bacillus cereus in raw milk and dairy products in Turkey. Int.J.Dairy Technol., 67(4): 562-569.
Hegde, R., Isloor, S., Prabhu, K.N., Shome, B.R., Rathnamma, D., Suryanarayana, V.V.S., Yatiraj, S., Prasad, C.R., Krishnaveni, N., Sundareshan, S. and Akhila, D.S. 2013. Incidence of subclinical mastitis and prevalence of major mastitis pathogens in organized farms and unorganized sectors. Indian J. Microbiol., 53(3): 315-320.
Hennekinne, J.A., DeBuyser, M.L. and Dragacci, S. 2012. Staphylococcus aureus and its food poisoning toxins: characterization and outbreak investigation. FEMS Microbiol. Rev., 36(4): 815-36.
Jørgensen, H.J., Mørk, T. and Rørvik, L.M. 2005. The occurrence of Staphylococcus aureus on a farm with small- scale production of raw milk cheese. J. Dairy. Sci., 88(11): 3810-3817.
Katsuda, K., Hata, E., Kobayashi, H., Kohmoto, M., Kawashima, K., Tsunemitsu, H. and Eguchi, M., 2005. Molecular typing of Staphylococcus aureus isolated from bovine mastitic milk on the basis of toxin genes and coagulase gene polymorphisms. Vet. Microbiol., 105(3-4): 301-305.
Köck, R., Becker, K., Cookson, B., van Gemert-Pijnen, J.E., Harbarth, S., Kluytmans, J.A.J.W., Mielke, M., Peters, G., Skov, R.L., Struelens, M.J. and Tacconelli, E. 2010. Methicillin-resistant Staphylococcus aureus (MRSA): burden of disease and control challenges in Europe. Euro. Surveill., 15(41): 19688.
Kreausukon, K., Fetsch, A., Kraushaar, B., Alt, K., Müller, K., Krömker, V., Zessin, K.H., Käsbohrer, A. and Tenhagen, B.A. 2012. Prevalence, antimicrobial resistance, and molecular characterization of methicillin-resistant Staphylococcus aureus from bulk tank milk of dairy herds. J. Dairy. Sci., 95(8): 4382-4388.
Kriegeskorte, A., Ballhausen, B., Idelevich, E.A., Köck, R., Friedrich, A.W., Karch, H., Peters, G. and Becker, K. 2012. Human MRSA isolates with novel genetic homolog, Germany. Emerg. Infect. Dis., 18(6): 1016.
Livermore, D.M. 2000. Antibiotic resistance in staphylococci. Int. J. Antimicrob. Agents, 16: 3-10.
Nemati, M., Hermans, K., Devriese, L.A., Maes, D. and Haesebrouck, F. 2009. Screening of genes encoding adhesion
factors and biofilm formation in Staphylococcus aureusisolates from poultry. Avian. Pathology, 38(6): 513-517.
Normanno, G., Corrente, M., La Salandra, G., Dambrosio, A., Quaglia, N.C., Parisi, A., Greco, G., Bellacicco, A.L., Virgilio, S. and Celano, G.V. 2007. Methicillin-resistant Staphylococcus aureus (MRSA) in foods of animal origin product in Italy. Int. J. Food Microbiol., 117(2): 219-222.
Riva, A., Borghi, E., Cirasola, D., Colmegna, S., Borgo, F., Amato, E., Pontello, M.M. and Morace, G. 2015. Methicillin- resistant Staphylococcus aureus in raw milk: Prevalence, SCC mec typing, enterotoxin characterization, and antimicrobial resistance patterns. J. Food Prot.,78(6): 1142-1146.
Ryffel, C. 1990. Sequence comparison of mecA genes isolated from methicillin-resistant Staphylococcus aureus and Staphylococcus epidermidis. Gene., 94: 137-138.
Sarkar, P., Mohanta, D. and Debnath, C. 2014. Staphylococcus aureus in dairy animals and farm workers in a closed herd in Karnal, North India: Assessment of prevalence rate and COA variations. Int. J. Innov. Res. Sci. Eng. Technol., 3(4): 10962-10972.
Sudhanthiramani, S., Swetha, C.S. and Bharathy, S. 2015. Prevalence of antibiotic resistant Staphylococcus aureusfrom raw milk samples collected from the local vendors in the region of Tirupathi, India. Vet. World, 8(4): 478.
Tong, S.Y., Davis, J.S., Eichenberger, E., Holland, T.L. and Fowler, V.G. 2015. Staphylococcus aureus infections: epidemiology, pathophysiology, clinical manifestations and management. Clin. Microbiol. Rev., 28(3): 603-661.
Veras, J.F., do Carmo, L.S., Tong, L.C., Shupp, J.W., Cummings, C., dos Santos, D.A., Cerqueira, M.M.O.P., Cantini, A., Nicoli, J.R. and Jett, M. 2008. A study of the enterotoxigenicity of coagulase-negative and coagulase-positive staphylococcal isolates from food poisoning outbreaks in Minas Gerais, Brazil. Int. J. Infect. Dis., 12(4): 410-415.